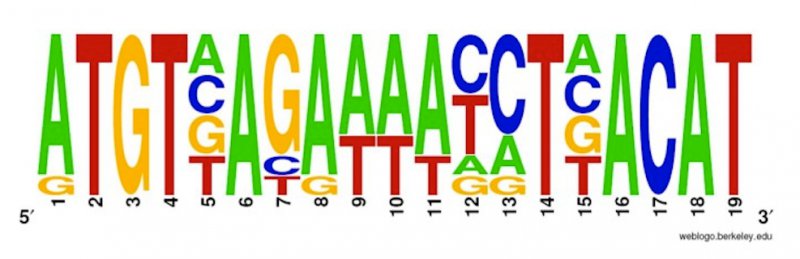
New algorithms are presented for the problem of multiple string matching of gapped patterns, where a gapped pattern is a sequence of strings such that there is a gap of fixed length between each two consecutive strings. The problem has applications in the discovery of transcription factor binding sites in DNA sequences when using generalized versions of the Position Weight Matrix model to describe transcription factor specificities. Existing algorithms are worst-case efficient but not practical, or vice versa, while the new algorithms lie in a middle-ground among the existing algorithms.
Emanuele Giaquinta, Kimmo Fredriksson, Szymon Grabowski, Alexandru I. Tomescu, Esko Ukkonen. Motif matching using gapped patterns. Theoretical Computer Science 548, 1-13, 2014
Last updated on 6 Oct 2014 by Aristides Gionis - Page created on 6 Oct 2014 by Aristides Gionis