An advanced computational model developed by computer scientists has revealed long delays in gene regulation.
The fields of genetics and genomics have developed rapidly during the past years, partly due to better computational methods. Now an international research group led by Finnish researchers has developed a new computational model, which has revealed unexpectedly long processing delays in gene expression following a regulatory signal.
The result will help us to understand life's basic processes in animals and plants and will form a foundation for developing more accurate algorithms to make better sense of gene regulation.
Method inside a method
The researchers studied the response of breast cancer cells to a hormonal regulatory signal that triggers changes in the expression of numerous genes. Some of the genes participate in the response through regulating other genes but these mechanisms are so far poorly understood. More accurate knowledge of gene regulation could enable for example the development of better cancer treatments.
– All gene regulation models are based on assumed effects of regulation, and, if the delay in gene regulation is not taken into account, these can lead to wrong conclusions. The importance of the delays may be compared to a situation where we want to investigate the cause for an allergy when some of the symptoms appear quickly, some only after a long delay. If attention is not paid to this, the investigation will be considerably more difficult and can produce wrong conclusions, Antti Honkela from the University of Helsinki points out.
– We built a physically inspired mathematical model within an advanced statistical model. This made it possible to observe, among other things, the delays of even over 20 minutes that we discovered in gene regulation, tells Jaakko Peltonen from Aalto University and University of Tampere.
New measurement technologies have enabled the investigation of biological phenomena on a vastly greater scale than before. Due to the growth in the scale of data, computational methods are central for all work in the fields of genetics and genomics.
The article Genome-wide modelling of transcription kinetics reveals patterns of RNA production delays was recently published in the Proceedings of the National Academy of Sciences of the United States of America.
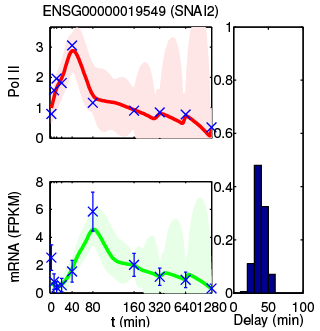
Figure: Examples of the observation data and fitted models for different genes. For each gene the upper red curve depicts its transcriptional activity and the lower green curve the expression of mature messenger RNA. The distribution of the delays estimated between these is shown on the right.
Contact person: Antti Honkela
Last updated on 9 Oct 2015 by Antti Honkela - Page created on 9 Oct 2015 by Antti Honkela