The combinatorial pattern-matching group develops combinatorial algorithms for pattern search and synthesis problems for sequential and higher-dimensional data. The group is interested in the basic research of the theoretical aspects of the area as well as in various applications, mostly in bioinformatics and information retrieval.
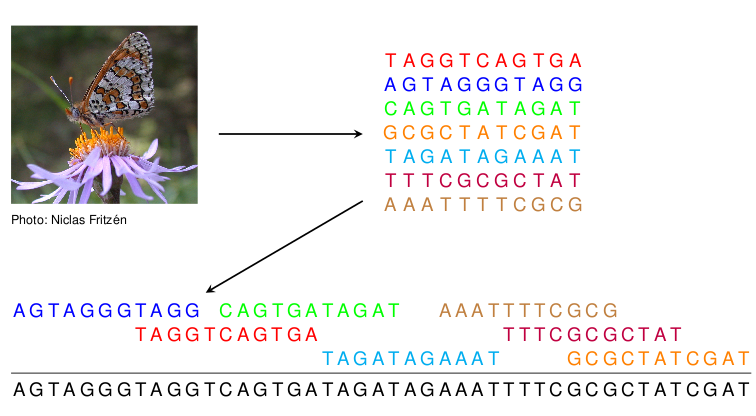
Recently we have focused in developing algorithms for assembling genomes from short read data. We have proposed new algorithms for correcting sequencing errors in the data. We have also developed new methods for the scaffolding phase where contiguous sequences assembled from the short read data are arranged into a linear order.
Another recent research focus is regulatory regions in genomes. We have developed new models based high throughput data for describing regulatory elements and we have proposed new algorithms for recognizing such elements in genomic sequences.
People:
Leader:
- Prof. Esko Ukkonen
Senior researchers and post-docs:
- Juha Kärkkäinen
- Leena Salmela
- Emanuele Giaquinta
- Simon Puglisi
- Kjell Lemström
PhD students:
- Jarkko Toivonen
- Teppo Ahonen
- Antti Laaksonen
- Dominik Kempa
Key publications:
- L. Salmela, V. Mäkinen, N. Välimäki, J. Ylinen, and E. Ukkonen: Fast scaffolding with small independent mixed integer programs. Bioinformatics 27(23):3259-3265, 2011.
- L. Salmela and J. Schröder: Correcting errors in short reads by multiple alignments. Bioinformatics 27(11):1455-1461, 2011. (Also in HiTSeq 2011).
- C. Pizzi, P. Rastas, and E. Ukkonen: Finding significant matches of position weight matrices in linear time. IEEE/ACM Transactions on Computational Biology and Bioinformatics 8(1):69-79, 2011.
- G.-H. Wei et al.: Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. The EMBO Journal 29:2147-2160, 2010.
Last updated on 5 Jun 2012 by Leena Salmela - Page created on 13 Jan 2007 by Webmaster